Scikit-learn
Table of contents:
- Loading Sample datasets from Scikit-learn
- Splitting the Dataset into training and test sets
- Preprocessing the Data for model fitting
- Machine learning algorithms
- Feature Engineering
- Resampling Methods
- Model evaluation
- Pipelines: Streamlining Machine Learning Workflows
- Model tuning
Scikit-learn is a Python library that provides a standard interface for implementing Machine Learning algorithms. It includes other ancillary functions that are integral to the machine learning pipeline such as data pre-processing steps, data resampling techniques, evaluation parameters and search interfaces for tuning/ optimizing an algorithms performance.
This section will go through the functions for implementing a typical machine learning pipeline with Scikit-learn. Since, Scikit-learn have a variety of packages and module that are called depending on the use-case, we’ll import a module directly from a package if and when needed using the from keyword. Again the goal of this material is to provide the foundation to be able to comb through the exhaustive Scikit-learn library and be able to use the right tool or function to get the job done.
The components covered include:
- Loading Sample datasets from Scikit-learn.
- Splitting the Dataset into training and test sets.
- Preprocessing the Data for model fitting.
- Initializing the machine learning estimator.
- Fitting the model.
- Model evaluation.
- Model tuning.
Loading Sample datasets from Scikit-learn
Scikit-learn comes with a set of small standard datasets for quickly testing and prototyping machine learning models. These dataset are ideal for learning purposes when starting off working with machine learning or even trying out the performance of some new model. They save a bit of the time required to identify, download and clean-up a dataset gotten from the wild. However, these datasets are small and well curated, they do not represent real-world scenarios.
Five popular sample datasets are:
- Boston house-prices dataset
- Diabetes dataset
- Iris dataset
- Wisconsin breast cancer dataset
- Wine dataset
The table below summarizes the properties of these datasets.
| Dataset name | Observations | Dimensions | Features | Targets |
|---|---|---|---|---|
| Boston house-prices dataset (regression) | 506 | 13 | real, positive | real 5. - 50. |
| Diabetes dataset (regression) | 442 | 10 | real, -.2 < x < .2 | integer 25 - 346 |
| Iris dataset (classification) | 150 | 4 | real, positive | 3 classes |
| Wisconsin breast cancer dataset (classification) | 569 | 30 | real, positive | 2 classes |
| Wine dataset (classification) | 178 | 13 | real, positive | 3 classes |
To load the sample dataset, we’ll run:
# load library
> from sklearn import datasets
Load the iris dataset
# load iris
> iris = datasets.load_iris()
> iris.data.shape
'Output': (150, 4)
iris.feature_names
'Output':
['sepal length (cm)',
'sepal width (cm)',
'petal length (cm)',
'petal width (cm)']
Methods for loading other datasets:
- Boston house-prices dataset -
datasets.load_boston() - Diabetes dataset -
datasets.load_diabetes() - Wisconsin breast cancer dataset -
datasets.load_breast_cancer() - Wine dataset -
datasets.load_wine()
Splitting the Dataset into training and test sets
A core practice in machine learning is to split the dataset into diffent partitions for training and testing. Scikit-learn has a convenient method to assist in that process called train_test_split(X, y, test_size=0.25), where X is the design matrix or dataset of predictors and y is the target variable. The split size is controlled using the attribute test_size. By default, test_size is set to 25% of the dataset size. It is standard practice to shuffle the dataset before splitting by setting the attribute shuffle=True.
# import module
> from sklearn.model_selection import train_test_split
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(iris.data, iris.target, shuffle=True)
> X_train.shape
'Output': (112, 4)
X_test.shape
'Output': (38, 4)
y_train.shape
'Output': (112,)
y_test.shape
'Output': (38,)
Preprocessing the Data for model fitting
Before a dataset is trained or fitted with a machine learning model, it necessarily undergoes some vital transformations. These transformation have a huge effect on the performance of the learning model. Transformations in Scikit-learn have a fit() and transform() method, or a fit_transform() method.
Depending on the use-case, the fit() method can be used to learn the parameters of the dataset, while the transform() method applies the data transform based on the learned parameters to the same dataset and also to the test or validation datasets before modeling. Also, the fit_transform() method can be used to learn and apply the transformation to the same dataset in a one-off fashion. Data transformation packages are found in the sklearn.preprocessing package.
This section will cover some critical transformation for numeric and categorical variables. They include:
- Data Rescaling
- Standardization
- Normalization
- Binarization
- Encoding Categorical Variables
- Input Missing Data
- Generating Higer Order Polynomial Features
Data Rescaling
It is often the case that the features of the dataset contain data with different scales. In other words, the data in column A can be in the range of 1 - 5, while the data in column B is in the range of 1000 - 9000. This different scales for units of observations in the same dataset can have an adverse effect for certain machine learning models, especially when minimizing the cost function of the algorithm because it shrinks the function space and makes it difficult for an optimization algorithm like gradient descent to find the global minimum.
When performing data rescaling, usually the attributes are rescaled with the range of 0 and 1. Data rescaling is implemted in Scikit-learn using the MinMaxScaler module. Let’s see an example.
# import packages
> from sklearn import datasets
> from sklearn.preprocessing import MinMaxScaler
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# print first 5 rows of X before rescaling
> X[0:5,:]
'Output':
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2]])
# rescale X
> scaler = MinMaxScaler(feature_range=(0, 1))
> rescaled_X = scaler.fit_transform(X)
# print first 5 rows of X after rescaling
> rescaled_X[0:5,:]
'Output':
array([[0.22222222, 0.625 , 0.06779661, 0.04166667],
[0.16666667, 0.41666667, 0.06779661, 0.04166667],
[0.11111111, 0.5 , 0.05084746, 0.04166667],
[0.08333333, 0.45833333, 0.08474576, 0.04166667],
[0.19444444, 0.66666667, 0.06779661, 0.04166667]])
Standardization
Linear Machine Learning algorithms such as Linear regression and Logistic regression make an assumption that the observations of the dataset are normally distributed with a mean of 0 and standard deviation of 1. However, this is often not the case with real-world datasets as features are often skewed with differing means and standard deviations.
By applying the technique of Standardization to the datasets transforms the features into a Standard Gaussian (or normal) distribution with a mean of 0 and standard deviation of 1. Scikit-learn implements data standardization in the StandardScaler module. Let’s see an example.
# import packages
> from sklearn import datasets
> from sklearn.preprocessing import StandardScaler
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# print first 5 rows of X before standardization
> X[0:5,:]
'Output':
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2]])
# standardize X
> scaler = StandardScaler().fit(X)
> standardize_X = scaler.transform(X)
# print first 5 rows of X after standardization
> standardize_X[0:5,:]
'Output':
array([[-0.90068117, 1.03205722, -1.3412724 , -1.31297673],
[-1.14301691, -0.1249576 , -1.3412724 , -1.31297673],
[-1.38535265, 0.33784833, -1.39813811, -1.31297673],
[-1.50652052, 0.10644536, -1.2844067 , -1.31297673],
[-1.02184904, 1.26346019, -1.3412724 , -1.31297673]])
Normalization
Data normalization involves transforming the observations in the dataset so that it has a unit norm or has magnitude or length of 1. The length of a vector is the square-root of the sum of squares of the vector elements. A unit vector (or unit norm) is obtained by dividing the vector by its length. Normalizing the dataset is particularly useful in scenarios where the dataset is sparse (i.e., a large number of observations are zeros) and also have differing scales. Normalization in Scikit-learn is implemented in the Normalizer module.
# import packages
> from sklearn import datasets
> from sklearn.preprocessing import Normalizer
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# print first 5 rows of X before normalization
> X[0:5,:]
'Output':
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2]])
# normalize X
> scaler = Normalizer().fit(X)
> normalize_X = scaler.transform(X)
# print first 5 rows of X after normalization
> normalize_X[0:5,:]
'Output':
array([[0.80377277, 0.55160877, 0.22064351, 0.0315205 ],
[0.82813287, 0.50702013, 0.23660939, 0.03380134],
[0.80533308, 0.54831188, 0.2227517 , 0.03426949],
[0.80003025, 0.53915082, 0.26087943, 0.03478392],
[0.790965 , 0.5694948 , 0.2214702 , 0.0316386 ]])
Binarization
Binarization is a transformation technique for converting a dataset into binary values by setting a cut-off or threshold. All values above the threshold are set to 1, while those below are set to 0. This technique is useful for converting a dataset of probabilities into integer values or in transforming a feature to reflect some categorization. Scikit-learn implements binarization with the Binarizer module.
# import packages
> from sklearn import datasets
> from sklearn.preprocessing import Binarizer
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# print first 5 rows of X before binarization
> X[0:5,:]
'Output':
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2]])
# binarize X
> scaler = Binarizer(threshold = 1.5).fit(X)
> binarize_X = scaler.transform(X)
# print first 5 rows of X after binarization
> binarize_X[0:5,:]
'Output':
array([[1., 1., 0., 0.],
[1., 1., 0., 0.],
[1., 1., 0., 0.],
[1., 1., 0., 0.],
[1., 1., 0., 0.]])
Encoding Categorical Variables
Most machine learing algorithms do not compute with non-numerical or categorical variables. Hence, encoding categorical variables is the technique for converting non-numerical features with labels into a numerical representation for use in machine learning modeling. Scikit-learn provides modules for encoding categorical variables including the LabelEncoder, for encoding labels as integers OneHotEncoder for converting categorical features into a matrix of integers (but first the categories will have to be encoded using LabelEncoder), and LabelBinarizer for creating a one hot encoding of target labels.
LabelEncoder is typically used on the target variable to transform a vector of hashable categories (or labels) into an integer representation by encoding label with values between 0 and the number of categories minus 1. This is further illustrated in Figure 1.
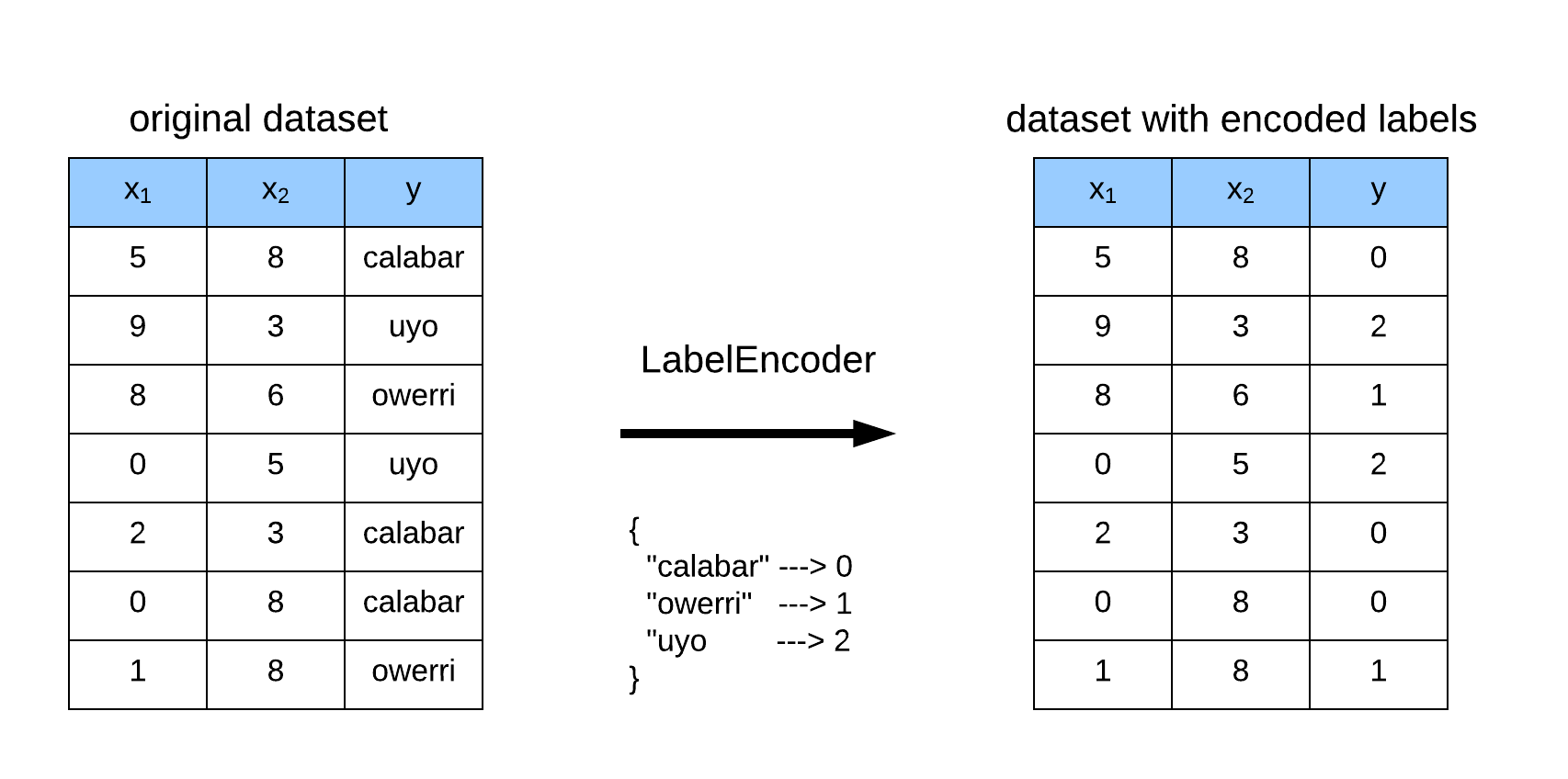
Let’s see an example of LabelEncoder
# import packages
> from sklearn.preprocessing import LabelEncoder
# create dataset
> data = np.array([[5,8,"calabar"],[9,3,"uyo"],[8,6,"owerri"],
[0,5,"uyo"],[2,3,"calabar"],[0,8,"calabar"],
[1,8,"owerri"]])
> data
'Output':
array([['5', '8', 'calabar'],
['9', '3', 'uyo'],
['8', '6', 'owerri'],
['0', '5', 'uyo'],
['2', '3', 'calabar'],
['0', '8', 'calabar'],
['1', '8', 'owerri']], dtype='<U21')
# separate features and target
> X = data[:,:2]
> y = data[:,-1]
# encode y
> encoder = LabelEncoder()
> encode_y = encoder.fit_transform(y)
# adjust dataset with encoded targets
> data[:,-1] = encode_y
> data
'Output':
array([['5', '8', '0'],
['9', '3', '2'],
['8', '6', '1'],
['0', '5', '2'],
['2', '3', '0'],
['0', '8', '0'],
['1', '8', '1']], dtype='<U21')
OneHotEncoder is used to transform a categorical feature variable in a matrix of integers. This matrix is a sparse matrix with each column corresponding to one possible value of a category. This is further illustrated in Figure 2.
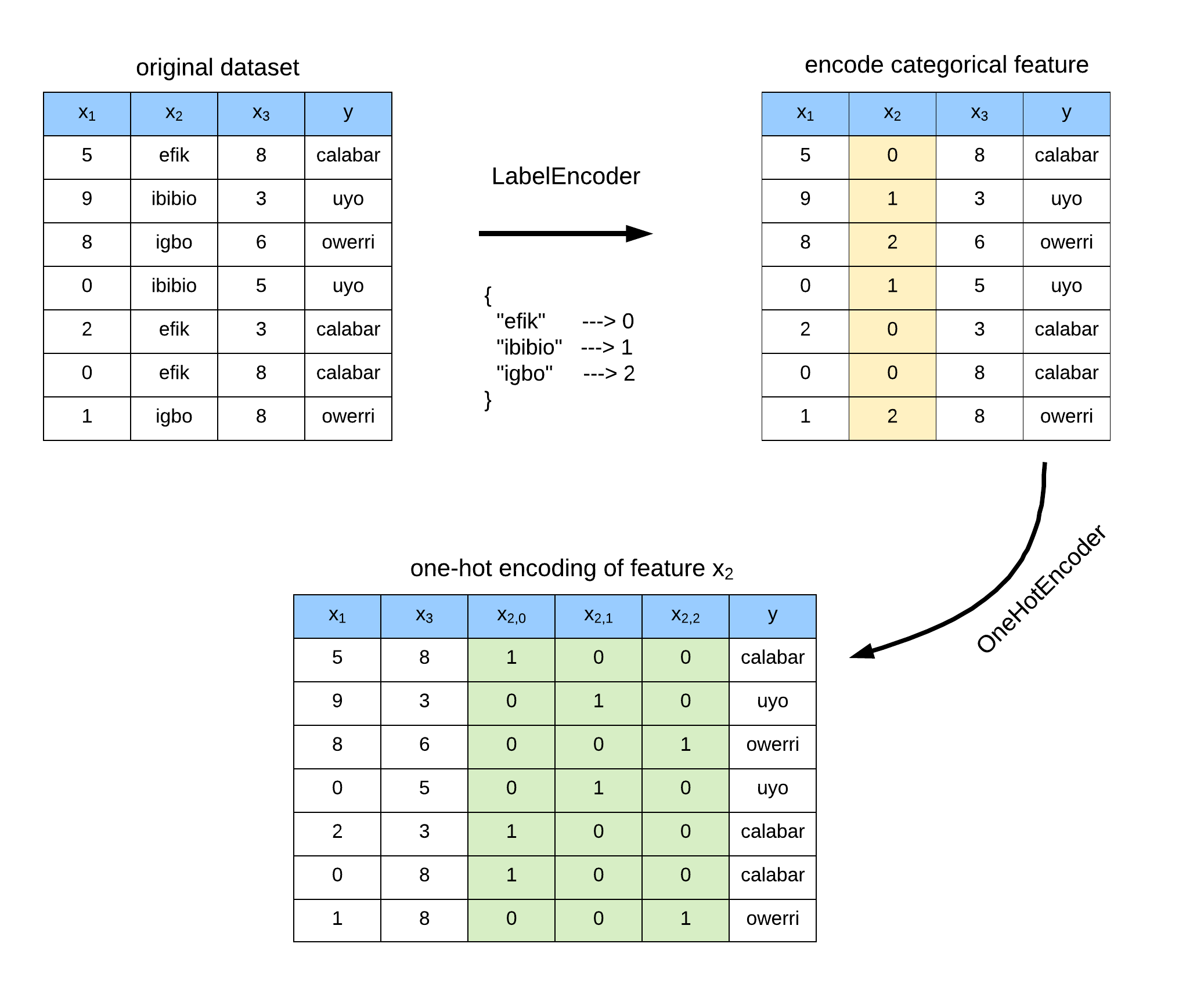
Let’s see an example of OneHotEncoder
# import packages
> from sklearn.preprocessing import LabelEncoder
> from sklearn.preprocessing import OneHotEncoder
# create dataset
> data = np.array([[5,"efik", 8,"calabar"],[9,"ibibio",3,"uyo"],[8,"igbo",6,"owerri"],
[0,"ibibio",5,"uyo"],[2,"efik",3,"calabar"],[0,"efik",8,"calabar"],
[1,"igbo",8,"owerri"]])
# separate features and target
> X = data[:,:3]
> y = data[:,-1]
# print the feature or design matrix X
> X
'Output':
array([['5', 'efik', '8'],
['9', 'ibibio', '3'],
['8', 'igbo', '6'],
['0', 'ibibio', '5'],
['2', 'efik', '3'],
['0', 'efik', '8'],
['1', 'igbo', '8']], dtype='<U21')
# label encode categorical features
> label_encoder = LabelEncoder()
> X[:,1] = label_encoder.fit_transform(data[:,1])
# print label encoding
> X[:,1]
'Output': array([0, 1, 2, 1, 0, 0, 2])
# print adjusted X
> X
'Output':
array([['5', '0', '8'],
['9', '1', '3'],
['8', '2', '6'],
['0', '1', '5'],
['2', '0', '3'],
['0', '0', '8'],
['1', '2', '8']], dtype='<U21')
# one_hot_encode X
> one_hot_encoder = OneHotEncoder()
> encode_categorical = X[:,1].reshape(len(X[:,1]), 1)
> one_hot_encode_X = one_hot_encoder.fit_transform(encode_categorical)
# print one_hot encoded matrix - use todense() to print sparse matrix
# or convert to array with toarray()
> one_hot_encode_X.todense()
'Output':
matrix([[1., 0., 0.],
[0., 1., 0.],
[0., 0., 1.],
[0., 1., 0.],
[1., 0., 0.],
[1., 0., 0.],
[0., 0., 1.]])
# remove categorical label
> X = np.delete(X, 1, axis=1)
# append encoded matrix
> X = np.append(X, one_hot_encode_X.toarray(), axis=1)
> X
'Output':
array([['5', '8', '1.0', '0.0', '0.0'],
['9', '3', '0.0', '1.0', '0.0'],
['8', '6', '0.0', '0.0', '1.0'],
['0', '5', '0.0', '1.0', '0.0'],
['2', '3', '1.0', '0.0', '0.0'],
['0', '8', '1.0', '0.0', '0.0'],
['1', '8', '0.0', '0.0', '1.0']], dtype='<U32')
Input Missing Data
It is often the case that a dataset contains several missing observations. Scikit-learn implements the Imputer module for completing missing values.
# import packages
> from sklearn.preprocessing import Imputer
# create dataset
> data = np.array([[5,np.nan,8],[9,3,5],[8,6,4],
[np.nan,5,2],[2,3,9],[np.nan,8,7],
[1,np.nan,5]])
> data
'Output':
array([[ 5., nan, 8.],
[ 9., 3., 5.],
[ 8., 6., 4.],
[nan, 5., 2.],
[ 2., 3., 9.],
[nan, 8., 7.],
[ 1., nan, 5.]])
# impute missing values - axix=0: impute along columns
> imputer = Imputer(missing_values='NaN', strategy='mean', axis=0)
> imputer.fit_transform(data)
'Output':
array([[5., 5., 8.],
[9., 3., 5.],
[8., 6., 4.],
[5., 5., 2.],
[2., 3., 9.],
[5., 8., 7.],
[1., 5., 5.]])
Generating Higer Order Polynomial Features
Sickit-learn has a module called PolynomialFeatures for generating a new dataset containing high-order polynomial and interaction features based off the features in the original dataset. For example, if the original dataset has two dimensions [a, b], the 2nd degree polynomial transformation of the features will result in [1, a, b, , ab, ].
# import packages
> from sklearn.preprocessing import PolynomialFeatures
# create dataset
> data = np.array([[5,8],[9,3],[8,6],
[5,2],[3,9],[8,7],
[1,5]])
> data
'Output':
array([[5, 8],
[9, 3],
[8, 6],
[5, 2],
[3, 9],
[8, 7],
[1, 5]])
# create polynomial features
> polynomial_features = PolynomialFeatures(2)
> data = polynomial_features.fit_transform(data)
> data
'Output':
array([[ 1., 5., 8., 25., 40., 64.],
[ 1., 9., 3., 81., 27., 9.],
[ 1., 8., 6., 64., 48., 36.],
[ 1., 5., 2., 25., 10., 4.],
[ 1., 3., 9., 9., 27., 81.],
[ 1., 8., 7., 64., 56., 49.],
[ 1., 1., 5., 1., 5., 25.]]
Machine learning algorithms
Scikit-learn provides convenient modules for implementing a variety of machine learning models. We’ll briefly survey implementing a few supervised and unsupervised machine learning models using Scikit-learn fantastic interface. Scikit-learn provides a consistent set of methods, which are the fit() method for fitting models to the training dataset and the predict() method for using the fitted parameters to make a prediction on the test dataset. The examples in this section is geared at explaining working with Scikit-learn, hence we are not so keen on the model performance.
Supervised learning
Linear Regression
# import packages
> from sklearn.linear_model import LinearRegression
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
# setting normalize to true normalizes the dataset before fitting the model
> linear_reg = LinearRegression(normalize = True)
# fit the model on the training set
> linear_reg.fit(X_train, y_train)
'Output': LinearRegression(copy_X=True, fit_intercept=True, n_jobs=1, normalize=True)
# make predictions on the test set
predictions = linear_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 14.46
Logistic Regression
# import packages
> from sklearn.linear_model import LogisticRegression
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> logistic_reg = LogisticRegression()
# fit the model on the training set
> logistic_reg.fit(X_train, y_train)
'Output':
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False)
# make predictions on the test set
predictions = logistic_reg.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 1.00
Support Vector Machines
SVC is the Support Vector Machine for Classification. SVR is the Support Vector Machine for Regression and LinearSVC is the Scalable Linear Support Vector Machine for classification implemented using liblinear. The default kernel for the SVC is the radial basis function (rbf).
# import packages
> from sklearn.svm import SVC
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> svc_model = SVC()
# fit the model on the training set
> svc_model.fit(X_train, y_train)
'Output':
SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma='auto', kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)
# make predictions on the test set
predictions = svc_model.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 0.95
Let’s use the SVM for Regression.
# import packages
> from sklearn.svm import SVR
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model, using the linear kernel
> svr_model = SVR(kernel="linear")
# fit the model on the training set
> svr_model.fit(X_train, y_train)
'Output':
SVR(C=1.0, cache_size=200, coef0=0.0, degree=3, epsilon=0.1, gamma='auto',
kernel='rbf', max_iter=-1, shrinking=True, tol=0.001, verbose=False)
# make predictions on the test set
predictions = svr_model.predict(X_test)
# evaluate the model performance using accuracy metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 33.34
Unsupervised learning
Unsupervised learning algorithms pass only the training data X to the fit() method.
K-Means Clustering
# import packages
> from sklearn.cluster import KMeans
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
# split in train and test sets
> X_train, X_test = train_test_split(X, shuffle=True)
# create the model. Since we know that the iris dataset has 3 classes, we set n_clusters = 3
> kmeans = KMeans(n_clusters=3, random_state=0)
# fit the model on the training set
> kmeans.fit(X_train)
'Output':
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=1, precompute_distances='auto',
random_state=0, tol=0.0001, verbose=0)
# print the clustered labels for the training points
> kmeans.labels_
'Output':
array([0, 2, 1, 0, 2, 1, 1, 0, 0, 0, 0, 1, 1, 0, 2, 0, 2, 1, 2, 0, 0, 1,
2, 0, 1, 1, 2, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 2, 1, 0, 2, 0, 2,
0, 2, 1, 2, 0, 2, 0, 0, 1, 0, 1, 1, 1, 1, 1, 2, 1, 1, 1, 2, 1, 0,
2, 2, 0, 1, 1, 1, 0, 0, 2, 1, 1, 0, 1, 1, 1, 2, 1, 0, 1, 2, 1, 1,
1, 2, 0, 0, 0, 2, 0, 1, 0, 0, 2, 2, 1, 2, 0, 1, 2, 1, 1, 2, 1, 1,
2, 1], dtype=int32)
# cluster the test set based on the KMeans model
> kmeans.predict(X_test)
'Output':
array([0, 1, 1, 0, 2, 2, 0, 0, 1, 1, 0, 1, 1, 2, 2, 1, 1, 0, 1, 2, 1, 2,
0, 0, 1, 2, 0, 2, 2, 1, 1, 2, 1, 0, 0, 0, 2, 1], dtype=int32)
Hierarchical Clustering
# import packages
> from sklearn.cluster import AgglomerativeClustering
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
# create the model. Since we know that the iris dataset has 3 classes, we set n_clusters = 3
> hierarchical = AgglomerativeClustering(n_clusters=3, linkage='ward')
# fit the model on the training set
> hierarchical.fit(X)
'Output':
AgglomerativeClustering(affinity='euclidean', compute_full_tree='auto',
connectivity=None, linkage='ward', memory=None, n_clusters=3,
pooling_func=<function mean at 0x105ef3488>)
# returns cluster labels.
> hierarchical.labels_
'Output':
array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 2, 0, 2, 2, 2, 2, 0, 2, 2, 2,
2, 2, 2, 0, 0, 2, 2, 2, 2, 0, 2, 0, 2, 0, 2, 2, 0, 0, 2, 2, 2, 2,
2, 0, 0, 2, 2, 2, 0, 2, 2, 2, 0, 2, 2, 2, 0, 2, 2, 0])
Ensemble Algorithms
Classification and Regression Trees
Let’s see an example of Classification and Regression Trees with the DecisionTreeClassifier for classification problems.
# import packages
> from sklearn.tree import DecisionTreeClassifier
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> tree_classifier = DecisionTreeClassifier()
# fit the model on the training set
> tree_classifier.fit(X_train, y_train)
'Output':
DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=None,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False, random_state=None,
splitter='best')
# make predictions on the test set
predictions = tree_classifier.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 0.95
Let’s see an example of Classification and Regression Trees with the DecisionTreeRegressor for regression problems.
# import packages
> from sklearn.tree import DecisionTreeRegressor
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> tree_reg = DecisionTreeRegressor()
# fit the model on the training set
> tree_reg.fit(X_train, y_train)
'Output':
DecisionTreeRegressor(criterion='mse', max_depth=None, max_features=None,
max_leaf_nodes=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
presort=False, random_state=None, splitter='best')
# make predictions on the test set
predictions = tree_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 26.55
Random Forests
Let’s see an example of Random Forests with the RandomForestClassifier for classification problems.
# import packages
> from sklearn.ensemble import RandomForestClassifier
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> rf_classifier = RandomForestClassifier()
# fit the model on the training set
> rf_classifier.fit(X_train, y_train)
'Output':
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=1,
oob_score=False, random_state=None, verbose=0,
warm_start=False)
# make predictions on the test set
predictions = rf_classifier.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 0.95
Let’s see an example of Random Forests with the RandomForestRegressor for regression problems.
# import packages
> from sklearn.ensemble import RandomForestRegressor
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> rf_reg = RandomForestRegressor()
# fit the model on the training set
> rf_reg.fit(X_train, y_train)
'Output':
RandomForestRegressor(bootstrap=True, criterion='mse', max_depth=None,
max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=1,
oob_score=False, random_state=None, verbose=0, warm_start=False)
# make predictions on the test set
predictions = rf_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 14.09
Gradient Boosting
Let’s see an example of Gradient Boosting with the GradientBoostingClassifier for classification problems.
# import packages
> from sklearn.ensemble import GradientBoostingClassifier
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> sgb_classifier = GradientBoostingClassifier()
# fit the model on the training set
> sgb_classifier.fit(X_train, y_train)
'Output':
GradientBoostingClassifier(criterion='friedman_mse', init=None,
learning_rate=0.1, loss='deviance', max_depth=3,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=100,
presort='auto', random_state=None, subsample=1.0, verbose=0,
warm_start=False)
# make predictions on the test set
predictions = sgb_classifier.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 0.95
Let’s see an example of Gradient Boosting with the GradientBoostingRegressor for regression problems.
# import packages
> from sklearn.ensemble import GradientBoostingRegressor
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> sgb_reg = GradientBoostingRegressor()
# fit the model on the training set
> sgb_reg.fit(X_train, y_train)
'Output':
GradientBoostingRegressor(alpha=0.9, criterion='friedman_mse', init=None,
learning_rate=0.1, loss='ls', max_depth=3, max_features=None,
max_leaf_nodes=None, min_impurity_decrease=0.0,
min_impurity_split=None, min_samples_leaf=1,
min_samples_split=2, min_weight_fraction_leaf=0.0,
n_estimators=100, presort='auto', random_state=None,
subsample=1.0, verbose=0, warm_start=False)
# make predictions on the test set
predictions = sgb_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 14.40
Extreme Gradient Boosting (XGBoost)
First we’ll install the XGBoost package on the Datalab instance by running:
> sudo pip install xgboost
Now, let’s see an example of Extreme Gradient Boosting (XGBoost) with the XGBClassifier for classification problems.
# import packages
> from xgboost import XGBClassifier
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> xgboost_classifier = XGBClassifier()
# fit the model on the training set
> xgboost_classifier.fit(X_train, y_train)
'Output':
XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bytree=1, gamma=0, learning_rate=0.1, max_delta_step=0,
max_depth=3, min_child_weight=1, missing=None, n_estimators=100,
n_jobs=1, nthread=None, objective='multi:softprob', random_state=0,
reg_alpha=0, reg_lambda=1, scale_pos_weight=1, seed=None,
silent=True, subsample=1)
# make predictions on the test set
predictions = xgboost_classifier.predict(X_test)
# evaluate the model performance using accuracy metric
print("Accuracy: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy: 0.89
Let’s see an example of Extreme Gradient Boosting (XGBoost) with the XGBRegressor for regression problems.
# import packages
> from xgboost import XGBRegressor
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> xgboost_reg = XGBRegressor()
# fit the model on the training set
> xgboost_reg.fit(X_train, y_train)
'Output':
XGBRegressor(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bytree=1, gamma=0, learning_rate=0.1, max_delta_step=0,
max_depth=3, min_child_weight=1, missing=None, n_estimators=100,
n_jobs=1, nthread=None, objective='reg:linear', random_state=0,
reg_alpha=0, reg_lambda=1, scale_pos_weight=1, seed=None,
silent=True, subsample=1)
# make predictions on the test set
predictions = xgboost_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 8.37
Feature Engineering
Feature engineering the the process of systematically choosing the set of features in the dataset that are useful and relevant to the learning problem. It is often the case that irrelevant features negatively affects the performance of the model. This section will review some techniques implemented in Scikit-learn for selecting relevant features from a dataset. The techniques surveyed include:
- Statistical tests to select the best $k$ features using the
SelectKBestmodule. - Recursive Feature Elimination (RFE) to recursively remove irrelevant features from the dataset.
- Principal Component Analysis to select the compnents that account for the variation in the dataset.
- Feature Imporances using Ensembled or Tree classifiers.
Statistical tests to select the best $k$ features using the SelectKBest module
The list below is a selection of statistical tests to use with SelectKBest. The choice depends on if the dataset target variable is numerical or categorical.
- ANOVA F-value,
f_classif(classification) - Chi-squared stats of non-negative features,
chi2(classification) - F-value,
f_regression(regression) - Mutual information for a continuous target,
mutual_info_regression(regression)
Let’s see an example using Chi-squared test to select the best variables.
# import packages
> from sklearn import datasets
> from sklearn.feature_selection import SelectKBest
> from sklearn.feature_selection import chi2
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# feature engineering. Let's see the best 3 features by setting k = 3
> kBest_chi = SelectKBest(score_func=chi2, k=3)
> fit_test = kBest_chi.fit(X, y)
# print test scores
> fit_test.scores_
'Output': array([ 10.81782088, 3.59449902, 116.16984746, 67.24482759])
From the test scores, the top 3 important features in the dataset are ranked from feature 3 -> 4 -> 1 and 2 in order. The data scientist can choose to drop the second column and observe the effect on the model performance.
We can transform the dataset to subset only the important features.
> adjusted_features = fit_test.transform(X)
> adjusted_features[0:5,:]
'Output':
array([[5.1, 1.4, 0.2],
[4.9, 1.4, 0.2],
[4.7, 1.3, 0.2],
[4.6, 1.5, 0.2],
[5. , 1.4, 0.2]])
The result drops the second column of the dataset.
Recursive Feature Elimination (RFE)
RFE is used together with a learning model to recursively select the desired number of top performing features.
Let’s use RFE with LinearRegression.
# import packages
> from sklearn.feature_selection import RFE
> from sklearn.linear_model import LinearRegression
> from sklearn import datasets
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# feature engineering
> linear_reg = LinearRegression()
> rfe = RFE(estimator=linear_reg, n_features_to_select=6)
> rfe_fit = rfe.fit(X, y)
# print the feature ranking
> rfe_fit.ranking_
'Output': array([3, 5, 4, 1, 1, 1, 8, 1, 2, 6, 1, 7, 1])
From the result, the 4th, 5th, 6th, 8th, 11th and 13th features are the top 6 features in the Boston dataset.
Principal Component Analysis
# import packages
> from sklearn.decomposition import PCA
> from sklearn import datasets
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
# create the model.
> pca = PCA(n_components=3)
# fit the model on the training set
> pca.fit(X)
'Output':
PCA(copy=True, iterated_power='auto', n_components=3, random_state=None,
svd_solver='auto', tol=0.0, whiten=False)
# examine the principal components percentage of variance explained
> pca.explained_variance_ratio_
'Output': array([0.92461621, 0.05301557, 0.01718514])
# print the principal components
> pca_dataset = pca.components_
> pca_dataset
'Output':
array([[ 0.36158968, -0.08226889, 0.85657211, 0.35884393],
[ 0.65653988, 0.72971237, -0.1757674 , -0.07470647],
[-0.58099728, 0.59641809, 0.07252408, 0.54906091]])
Feature Imporances
Tree based or ensemble methods in Scikit-learn have a feature_importances_ attribute which can be used to drop irrelevant features in the dataset using the SelectFromModel module contained in the sklearn.feature_selection package.
Let’s used the Ensemble metod AdaBoostClassifier in this example.
# import packages
> from sklearn.ensemble import AdaBoostClassifier
> from sklearn.feature_selection import SelectFromModel
> from sklearn import datasets
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# feature engineering
> ada_boost_classifier = AdaBoostClassifier()
> ada_boost_classifier.fit(X, y)
'Output':
AdaBoostClassifier(algorithm='SAMME.R', base_estimator=None,
learning_rate=1.0, n_estimators=50, random_state=None)
# print the feature importances
> ada_boost_classifier.feature_importances_
'Output': array([0. , 0. , 0.58, 0.42])
# create a subset of data based on the relevant features
> model = SelectFromModel(ada_boost_classifier, prefit=True)
> new_data = model.transform(X)
# the irrelevant features have been removed
> new_data.shape
'Output': (150, 2)
Resampling Methods
Resampling methods are a set of techniques that involves selecting a subset of the available dataset, training on that data subset and using the reminder of the data to evaluate the trained model. Let’s review the techniques for resampling using Scikit-learn. This section covers:
- k-Fold cross validation, and
- Leave-One-Out cross validation
k-Fold cross validation
In k-Fold cross validation, the dataset is divided into k-parts or folds. The model is trained using folds, and evaluated on the remaining fold. This process is repeated k-times so that each fold can serve as a test set. At the end of the process, k-Fold averages the result and reports a mean score with a standard deviation. Scikit-learn implements K-Fold CV in the module KFold. The module cross_val_score is used to evaluate the cross-validation score using the splitting strategy, which is KFold in this case.
Let’s see an example of this using the k-Nearest Neighbors (kNN) classification algorithm. When initializing KFold it is standard practice to shuffle the data before splitting.
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
> from sklearn.neighbors import KNeighborsClassifier
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# initialize KFold - with shuffle = True, shuffle the data before splitting
> kfold = KFold(n_splits=3, shuffle=True)
# create the model
> knn_clf = KNeighborsClassifier(n_neighbors=3)
# fit the model using cross validation
> cv_result = cross_val_score(knn_clf, X, y, cv=kfold)
# evaluate the model performance using accuracy metric
print("Accuracy: %.3f%% (%.3f%%)" % (cv_result.mean()*100.0, cv_result.std()*100.0))
'Output':
Accuracy: 93.333% (2.494%)
Leave-One-Out cross validation (LOOCV)
In LOOCV just one example is assigned to the test set, and the model is trained on the remainder of the dataset. This process is repeated for all the examples in the dataset. This process is repeated until all the examples in the dataset have been used for evaluating the model.
> from sklearn.model_selection import LeaveOneOut
> from sklearn.model_selection import cross_val_score
> from sklearn.neighbors import KNeighborsClassifier
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# initialize LOOCV
> loocv = LeaveOneOut()
# create the model
> knn_clf = KNeighborsClassifier(n_neighbors=3)
# fit the model using cross validation
> cv_result = cross_val_score(knn_clf, X, y, cv=loocv)
# evaluate the model performance using accuracy metric
print("Accuracy: %.3f%% (%.3f%%)" % (cv_result.mean()*100.0, cv_result.std()*100.0))
'Output':
Accuracy: 96.000% (19.596%)
Model evaluation
This chapter has already used a couple of evaluation metrics for accessing the quality of the fitted models. In this section, we survey a couple of other metrics for regression and classification use-cases and how to implement them using Scikit-learn. For each metric, we show how to use them as standalone implementations, as well as together with cross validation using the cross_val_score method.
What we’ll cover here includes:
- Regression evaluation metrics
- Mean Squared Error (MSE): the average sum of squared difference between the predicted label, and the true label, . A score of 0 indicates a perfect prediction without errors.
- Mean Absolute Error (MAE): the average absolute differece between the predicted label, and the true label, . A score of 0 indicates a perfect prediction without errors.
- : the amount of variance or variability in the dataset explained by the model. The score of 1 means that the model perfectly captures the variability in the dataset.
- Classification evaluation metrics
- Accuracy: is the ratio of correct predictions to the total number of predictions. The bigger the accuracy, the better the model.
- Logarithmic Loss (aka logistic loss or cross-entropy loss): is the probability that an observation is correctly assigned to a class label. By minimizing the log-loss, conversely, the accuracy is maximized. So with this metric, values closer to zero are good.
- Area under the ROC curve (AUC-ROC): used in the binary classification case. Implemtation not provided, but very similar in style to the others.
- Confusion Matrix: more intuitive in the binary classification case. Implemtation not provided, but very similar in style to the others.
- Classification Report: it returns a text report of the main classification metrics.
Regression evaluation metrics
An example of regression evaluation metrics implemented stand-alone.
# import packages
> from sklearn.linear_model import LinearRegression
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import mean_squared_error
> from sklearn.metrics import mean_absolute_error
> from sklearn.metrics import r2_score
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
# setting normalize to true normalizes the dataset before fitting the model
> linear_reg = LinearRegression(normalize = True)
# fit the model on the training set
> linear_reg.fit(X_train, y_train)
'Output': LinearRegression(copy_X=True, fit_intercept=True, n_jobs=1, normalize=True)
# make predictions on the test set
predictions = linear_reg.predict(X_test)
# evaluate the model performance using mean square error metric
print("Mean squared error: %.2f" % mean_squared_error(y_test, predictions))
'Output':
Mean squared error: 14.46
# evaluate the model performance using mean absolute error metric
print("Mean absolute error: %.2f" % mean_absolute_error(y_test, predictions))
'Output':
Mean absolute error: 3.63
# evaluate the model performance using r-squared error metric
print("R-squared score: %.2f" % r2_score(y_test, predictions))
'Output':
R-squared score: 0.69
An example of regression evaluation metrics implemented with cross validation. Evaluation metrics for MSE and MAE using cross validation, MSE are implemented with the sign inverted. The simple way to interpret this is to have it in mind that the closer the values are to zero, the better the model.
> from sklearn.linear_model import LinearRegression
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# initialize KFold - with shuffle = True, shuffle the data before splitting
> kfold = KFold(n_splits=3, shuffle=True)
# create the model
> linear_reg = LinearRegression(normalize = True)
# fit the model using cross validation - score with Mean square error (MSE)
> mse_cv_result = cross_val_score(linear_reg, X, y, cv=kfold, scoring="neg_mean_squared_error")
# print mse cross validation output
print("Negtive Mean squared error: %.3f%% (%.3f%%)" % (mse_cv_result.mean(), mse_cv_result.std()))
'Output':
Negtive Mean squared error: -24.275% (4.093%)
# fit the model using cross validation - score with Mean absolute error (MAE)
> mae_cv_result = cross_val_score(linear_reg, X, y, cv=kfold, scoring="neg_mean_absolute_error")
# print mse cross validation output
print("Negtive Mean absolute error: %.3f%% (%.3f%%)" % (mae_cv_result.mean(), mse_cv_result.std()))
'Output':
Negtive Mean absolute error: -3.442% (4.093%)
# fit the model using cross validation - score with R-squared
> r2_cv_result = cross_val_score(linear_reg, X, y, cv=kfold, scoring="r2")
# print mse cross validation output
print("R-squared score: %.3f%% (%.3f%%)" % (r2_cv_result.mean(), r2_cv_result.std()))
'Output':
R-squared score: 0.707% (0.030%)
Classification evaluation metrics
An example of classification evaluation metrics implemented stand-alone.
# import packages
> from sklearn.linear_model import LogisticRegression
> from sklearn import datasets
> from sklearn.model_selection import train_test_split
> from sklearn.metrics import accuracy_score
> from sklearn.metrics import log_loss
> from sklearn.metrics import classification_report
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# split in train and test sets
> X_train, X_test, y_train, y_test = train_test_split(X, y, shuffle=True)
# create the model
> logistic_reg = LogisticRegression()
# fit the model on the training set
> logistic_reg.fit(X_train, y_train)
'Output':
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False)
# make predictions on the test set
predictions = logistic_reg.predict(X_test)
# evaluate the model performance using accuracy
print("Accuracy score: %.2f" % accuracy_score(y_test, predictions))
'Output':
Accuracy score: 0.89
# evaluate the model performance using log loss
### output the probabilities of assigning an observation to a class
predictions_probabilities = logistic_reg.predict_proba(X_test)
print("Log-Loss likelihood: %.2f" % log_loss(y_test, predictions_probabilities))
'Output':
Log-Loss likelihood: 0.39
# evaluate the model performance using classification report
print("Classification report: \n", classification_report(y_test, predictions, target_names=data.target_names))
'Output':
Classification report:
precision recall f1-score support
setosa 1.00 1.00 1.00 12
versicolor 0.85 0.85 0.85 13
virginica 0.85 0.85 0.85 13
avg / total 0.89 0.89 0.89 38
Let’s see an example of classification evaluation metrics implemented with cross validation. Evaluation metrics for Log-Loss using cross validation is implemented with the sign inverted. The simple way to interpret this is to have it in mind that the closer the values are to zero, the better the model.
> from sklearn.linear_model import LogisticRegression
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# initialize KFold - with shuffle = True, shuffle the data before splitting
> kfold = KFold(n_splits=3, shuffle=True)
# create the model
> logistic_reg = LogisticRegression()
# fit the model using cross validation - score with accuracy
> accuracy_cv_result = cross_val_score(logistic_reg, X, y, cv=kfold, scoring="accuracy")
# print accuracy cross validation output
print("Accuracy: %.3f%% (%.3f%%)" % (accuracy_cv_result.mean(), accuracy_cv_result.std()))
'Output':
Accuracy: 0.953% (0.025%)
# fit the model using cross validation - score with Log-Loss
> logloss_cv_result = cross_val_score(logistic_reg, X, y, cv=kfold, scoring="neg_log_loss")
# print mse cross validation output
print("Log-Loss likelihood: %.3f%% (%.3f%%)" % (logloss_cv_result.mean(), logloss_cv_result.std()))
'Output':
Log-Loss likelihood: -0.348% (0.027%)
Pipelines: Streamlining Machine Learning Workflows
The concept of pipelines in scikit-learn is a compelling tool for chaining a bunch of operations together to form a tidy process flow of data transforms from one state to another. The operations that constitute a pipeline can be any of scikit-learn’s transformers (i.e., modules with a fit and transform method, or a fit_transform method) or classifiers (i.e., modules with a fit and predict method, or a fit_predict method). Classifiers are also called predictors.
For a typical machine learning workflow, the steps taken may involve cleaning the data, feature engineering, scaling the dataset and then fitting a model. Pipelines can be used in this case to chain these operations together into a coherent workflow. They have the advantage of providing a convenient and consistent interface for calling at once a sequence of operations.
These transformers or predictors are collectively called estimators in scikit-learn terminology. In the last two paragraphs, we called them operations.
Another advantage of pipelines is that it safeguards against accidentally fitting a transform on the entire dataset and thereby leaking statistics influenced by the test data to the machine learning model while training. For example, if a standardizer is fitted on the whole dataset, the test set will be compromised because the test observations have contributed in estimating the mean and standard deviation for scaling the training set before fitting the model.
Finally, only the last step of the pipeline can be a classifier or predictor. All the stages of the pipeline must contain a transform method except the final stage, which can be a transformer or a classifier.
To begin using scikit-learn pipelines, first import:
from sklearn.pipeline import Pipeline
Let’s see some examples of working with Pipelines in Scikit-learn. In the following example, we’ll apply a scaling transform to standardize our dataset and then use a Support Vector Classifier to train the model.
# import packages
> from sklearn.svm import SVC
> from sklearn import datasets
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
> from sklearn.pipeline import Pipeline
# load dataset
> data = datasets.load_iris()
# separate features and target
> X = data.data
> y = data.target
# create the pipeline
> estimators = [
('standardize' , StandardScaler()),
('svc', SVC())
]
# build the pipeline model
> pipe = Pipeline(estimators)
# run the pipeline
> kfold = KFold(n_splits=3, shuffle=True)
> cv_result = cross_val_score(pipe, X, y, cv=kfold)
# evaluate the model performance
> print("Accuracy: %.3f%% (%.3f%%)" % (cv_result.mean()*100.0, cv_result.std()*100.0))
'Output':
Accuracy: 94.667% (0.943%)
Pipelines using make_pipeline
Another method for building machine learning pipelines is by using the make_pipeline method. For the next example, we use PCA to select the best six features and reduce the dimensionality of the dataset, and then we’ll fit the model using Random Forests for regression.
> from sklearn.pipeline import make_pipeline
> from sklearn.svm import SVR
> from sklearn import datasets
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
> from sklearn.decomposition import PCA
> from sklearn.pipeline import Pipeline
> from sklearn.ensemble import RandomForestRegressor
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# build the pipeline model
> pipe = make_pipeline(
PCA(n_components=9),
RandomForestRegressor()
)
# run the pipeline
> kfold = KFold(n_splits=4, shuffle=True)
> cv_result = cross_val_score(pipe, X, y, cv=kfold)
# evaluate the model performance
> print("Accuracy: %.3f%% (%.3f%%)" % (cv_result.mean()*100.0, cv_result.std()*100.0))
'Output':
Accuracy: 73.750% (2.489%)
Pipelines using FeatureUnion
Scikit-learn provides a module for merging the output of several transformers called feature_union. It does this by fitting each transformer independently to the dataset, and then their respective outputs are combined to form a transformed dataset for training the model.
FeatureUnion works in the same way as a Pipeline, and in many ways can be thought of as a means of building complex pipelines within a Pipeline.
Let’s see an example using FeatureUnion. Here, we will combine the output of Recursive Feature Elimination (RFE) and PCA for feature engineering, and then we’ll apply the Stochastic Gradient Boosting (SGB) ensemble model for regression to train the model.
> from sklearn.ensemble import GradientBoostingRegressor
> from sklearn.ensemble import RandomForestRegressor
> from sklearn import datasets
> from sklearn.model_selection import KFold
> from sklearn.model_selection import cross_val_score
> from sklearn.feature_selection import RFE
> from sklearn.decomposition import PCA
> from sklearn.pipeline import Pipeline
> from sklearn.pipeline import make_pipeline
> from sklearn.pipeline import make_union
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# construct pipeline for feature engineering - make_union similar to make_pipeline
> feature_engr = make_union(
RFE(estimator=RandomForestRegressor(), n_features_to_select=6),
PCA(n_components=9)
)
# build the pipeline model
> pipe = make_pipeline(
feature_engr,
GradientBoostingRegressor()
)
# run the pipeline
> kfold = KFold(n_splits=4, shuffle=True)
> cv_result = cross_val_score(pipe, X, y, cv=kfold)
# evaluate the model performance
> print("Accuracy: %.3f%% (%.3f%%)" % (cv_result.mean()*100.0, cv_result.std()*100.0))
'Output':
Accuracy: 88.956% (1.493%)
Model tuning
Each machine learning model has a set of options or configurations that can be tuned to optimize the model when fitting to data. This configurations are called hyper-parameters. Hence, for each hyper-parameter there exist a range of values that can be chosen. Taking into consideration the number of hyper-parameters that an algorithm has, the entire space can become exponentially large and infeasibile to explore all of them. Scikit-learn provides two convenient modules for searching through the hyper-parameters space of an agorithm to find the best values for each hyper-parameters that optimizes the model.
These modules are the:
- Grid Search, and
- Randomized Search
Grid Search
Grid search comprehensively explores all the specified hyper-parameter values for an estimator. It is implemented using the GridSearchCV module. Let’s see an example using the Random Forest for Regression. The hyper-parameters we’ll search over are:
- the number of trees in the forest,
n_estimators, - the maximum depth of the tree,
max_depth, and - the minimum number of samples required to split an internal node,
min_samples_leaf.
> from sklearn.model_selection import GridSearchCV
> from sklearn.ensemble import RandomForestRegressor
> from sklearn import datasets
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# construct grid search parameters in a dictionary
> parameters = {
'n_estimators': [2, 4, 6, 8, 10, 12, 14, 16],
'max_depth': [2, 4, 6, 8],
'min_samples_leaf': [1,2,3,4,5]
}
# create the model
> rf_model = RandomForestRegressor()
# run the grid search
> grid_search = GridSearchCV(estimator=rf_model, param_grid=parameters)
# fit the model
> grid_search.fit(X,y)
'Output':
GridSearchCV(cv=None, error_score='raise',
estimator=RandomForestRegressor(bootstrap=True, criterion='mse', max_depth=None,
max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=1,
oob_score=False, random_state=None, verbose=0, warm_start=False),
fit_params=None, iid=True, n_jobs=1,
param_grid={'n_estimators': [2, 4, 6, 8, 10, 12, 14, 16], 'max_depth': [2, 4, 6, 8], 'min_samples_leaf': [1, 2, 3, 4, 5]},
pre_dispatch='2*n_jobs', refit=True, return_train_score='warn',
scoring=None, verbose=0)
# evaluate the model performance
> print("Best Accuracy: %.3f%%" % (grid_search.best_score_*100.0))
'Output':
Best Accuracy: 57.917%
# best set of hyper-parameter values
> print("Best n_estimators: %d \nBest max_depth: %d \nBest min_samples_leaf: %d " % \
(grid_search.best_estimator_.n_estimators, \
grid_search.best_estimator_.max_depth, \
grid_search.best_estimator_.min_samples_leaf))
'Output':
Best n_estimators: 14
Best max_depth: 8
Best min_samples_leaf: 1
Randomized Search
As opposed to Grid Search, not all the provided hyper-parameter values are evaluated, but rather a determined number of hyper-parameter values are sampled from a random uniform distribution. The number of hyper-parameter values that can be evaluated is determined by the n_iter attribute of the RandomizedSearchCV module.
In this example, we will use the same scenario as in the Grid Search case.
> from sklearn.model_selection import RandomizedSearchCV
> from sklearn.ensemble import RandomForestRegressor
> from sklearn import datasets
# load dataset
> data = datasets.load_boston()
# separate features and target
> X = data.data
> y = data.target
# construct grid search parameters in a dictionary
> parameters = {
'n_estimators': [2, 4, 6, 8, 10, 12, 14, 16],
'max_depth': [2, 4, 6, 8],
'min_samples_leaf': [1,2,3,4,5]
}
# create the model
> rf_model = RandomForestRegressor()
# run the grid search
> randomized_search = RandomizedSearchCV(estimator=rf_model, param_distributions=parameters, n_iter=10)
# fit the model
> randomized_search.fit(X,y)
'Output':
RandomizedSearchCV(cv=None, error_score='raise',
estimator=RandomForestRegressor(bootstrap=True, criterion='mse', max_depth=None,
max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=10, n_jobs=1,
oob_score=False, random_state=None, verbose=0, warm_start=False),
fit_params=None, iid=True, n_iter=10, n_jobs=1,
param_distributions={'n_estimators': [2, 4, 6, 8, 10, 12, 14, 16], 'max_depth': [2, 4, 6, 8], 'min_samples_leaf': [1, 2, 3, 4, 5]},
pre_dispatch='2*n_jobs', random_state=None, refit=True,
return_train_score='warn', scoring=None, verbose=0)
# evaluate the model performance
> print("Best Accuracy: %.3f%%" % (randomized_search.best_score_*100.0))
'Output':
Best Accuracy: 57.856%
# best set of hyper-parameter values
> print("Best n_estimators: %d \nBest max_depth: %d \nBest min_samples_leaf: %d " % \
(randomized_search.best_estimator_.n_estimators, \
randomized_search.best_estimator_.max_depth, \
randomized_search.best_estimator_.min_samples_leaf))
'Output':
Best n_estimators: 12
Best max_depth: 6
Best min_samples_leaf: 5